While using Pandas in Python, it's common to filter data with df.query() method. But there are so many options of the syntax and parameters! In this tutorial, we will try to show the most important ones.

Simple Check
One of the simplest ways to filter data is to use a comparison operator:
df = pandas.read_csv('healthdata.csv') df2 = df.query("Gender == 'Male'")This will return all rows where the Gender column is equal to Male:
Person ID Gender Age ... Heart Rate Daily Steps Sleep Disorder0 1 Male 27.0 ... 77 4200 NaN1 2 Male 28.0 ... 75 10000 NaN2 3 Male 28.0 ... 75 10000 NaN3 4 Male 28.0 ... 85 3000 Sleep Apnea4 5 Male 28.0 ... 85 3000 Sleep Apnea [5 rows x 13 columns]Checking Against Variables
In case you have a dynamic value, you can use a variable in the query:
df = pandas.read_csv('healthdata.csv') value = 'Sleep Apnea'df2 = df.query("`Sleep Disorder` == @value")This will return all rows where the Sleep Disorder column is equal to the value of the value variable:
Person ID Gender Age ... Heart Rate Daily Steps Sleep Disorder3 4 Male 28.0 ... 85 3000 Sleep Apnea4 5 Male 28.0 ... 85 3000 Sleep Apnea16 17 Female 29.0 ... 80 4000 Sleep Apnea17 18 Male 29.0 ... 70 8000 Sleep Apnea30 31 Female 30.0 ... 78 4100 Sleep Apnea [5 rows x 13 columns]Fields With Spaces
Now, if your field has a space in the name, like in our previous example - you can use backticks to escape the field name:
df = pandas.read_csv('healthdata.csv') df2 = df.query("`Sleep Disorder` == 'Sleep Apnea'")This is because without the backticks, the query would fail:
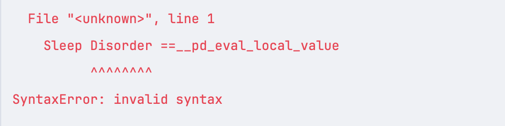
Multiple Conditions
Next, let's see how we can use multiple conditions:
df = pandas.read_csv('healthdata.csv') df2 = df.query("Gender == 'Male' and `Age` >= 29")This has allowed us to filter the data to only include rows where the Gender is Male and the Age is greater than or equal to 29:
Person ID Gender Age ... Heart Rate Daily Steps Sleep Disorder6 7 Male 29.0 ... 82 3500 Insomnia7 8 Male 29.0 ... 70 8000 NaN8 9 Male 29.0 ... 70 8000 NaN9 10 Male 29.0 ... 70 8000 NaN10 11 Male 29.0 ... 70 8000 NaN [5 rows x 13 columns]Of course, you can use or as well:
df2 = df.query("`Heart Rate` >= 80 or `Age` <= 27")This gives us all rows where the Heart Rate is greater than or equal to 80 or the Age is less than or equal to 27:
Person ID Gender Age ... Heart Rate Daily Steps Sleep Disorder0 1 Male 27.0 ... 77 4200 NaN3 4 Male 28.0 ... 85 3000 Sleep Apnea4 5 Male 28.0 ... 85 3000 Sleep Apnea5 6 Male 28.0 ... 85 3000 Insomnia6 7 Male 29.0 ... 82 3500 Insomnia [5 rows x 13 columns]Multiple Conditions Faster
But what if we want to use a range condition on a column? We can write it like this:
df = pandas.read_csv('healthdata.csv') df2 = df.query("30 <= Age and Age < 50")It filters our entries:
Person ID Gender Age ... Heart Rate Daily Steps Sleep Disorder19 20 Male 30.0 ... 70 8000 NaN20 21 Male 30.0 ... 70 8000 NaN21 22 Male 30.0 ... 70 8000 NaN22 23 Male 30.0 ... 70 8000 NaN23 24 Male 30.0 ... 70 8000 NaN [5 rows x 13 columns]But there's a better way to write this - combining the conditions into a single one:
df = pandas.read_csv('healthdata.csv') df2 = df.query('30 <= Age < 50')This will give us the same result but with more readable code.
Using the in Operator
Sometimes, you want to filter your data based on a list of values. You can do this using the in operator:
df = pandas.read_csv('healthdata.csv') df2 = df.query("`Sleep Disorder` in ['Sleep Apnea', 'Insomnia']")This will give us all rows where the Sleep Disorder column is either Sleep Apnea or Insomnia:
Person ID Gender Age ... Heart Rate Daily Steps Sleep Disorder3 4 Male 28.0 ... 85 3000 Sleep Apnea4 5 Male 28.0 ... 85 3000 Sleep Apnea5 6 Male 28.0 ... 85 3000 Insomnia6 7 Male 29.0 ... 82 3500 Insomnia16 17 Female 29.0 ... 80 4000 Sleep Apnea [5 rows x 13 columns]This also supports variables and not in operator:
list = ['Sleep Apnea', 'Insomnia']df2 = df.query("`Sleep Disorder` not in @list")It's a great way to build some logic into your queries.
Using F-Strings
Another way to use variables in queries is to use f-strings:
df = pandas.read_csv('healthdata.csv') value = 30df2 = df.query(f'Age < {value}')This does the same thing as before, but you don't need to add the @ symbol:
Person ID Gender Age ... Heart Rate Daily Steps Sleep Disorder0 1 Male 27.0 ... 77 4200 NaN1 2 Male 28.0 ... 75 10000 NaN2 3 Male 28.0 ... 75 10000 NaN3 4 Male 28.0 ... 85 3000 Sleep Apnea4 5 Male 28.0 ... 85 3000 Sleep Apnea [5 rows x 13 columns]Math Actions
Next, we want to show you how to use math in queries. Let's say we want to filter our data based on a math operation:
df = pandas.read_csv('healthdata.csv') df2 = df.query('Age <= `Heart Rate` / 3')This compares our Age column to the result of the Heart Rate column divided by 3:
Person ID Gender Age ... Heart Rate Daily Steps Sleep Disorder3 4 Male 28.0 ... 85 3000 Sleep Apnea4 5 Male 28.0 ... 85 3000 Sleep Apnea5 6 Male 28.0 ... 85 3000 Insomnia [3 rows x 13 columns]We can also write this as multiplication:
df2 = df.query('Age <= `Heart Rate` * 0.33')And it will still give us the same result.
String Method Matches
Our following example involves string methods. Let's say we want to filter our data based on a string method and only take rows where the Blood Pressure column ends with /80:
df = pandas.read_csv('healthdata.csv') # String methods:df2 = df.query('`Blood Pressure`.str.endswith("/80")')This will give us all rows where the Blood Pressure column ends with /80:
Person ID Gender Age Occupation Sleep Duration Quality of Sleep \1 2 Male 28.0 Doctor 6.2 62 3 Male 28.0 Doctor 6.2 67 8 Male 29.0 Doctor 7.8 78 9 Male 29.0 Doctor 7.8 79 10 Male 29.0 Doctor 7.8 7 Physical Activity Level Stress Level BMI Category Blood Pressure \1 60 8 Normal 125/802 60 8 Normal 125/807 75 6 Normal 120/808 75 6 Normal 120/809 75 6 Normal 120/80 Heart Rate Daily Steps Sleep Disorder1 75 10000 NaN2 75 10000 NaN7 70 8000 NaN8 70 8000 NaN9 70 8000 NaNNote: We have used pandas.set_option('display.max_columns', None) to display all columns in the output.
This can be very useful when you filter your data based on some part of strings or apply any other string method.
Take Non-Null Values
Pandas also allows you to filter by not null values:
df = pandas.read_csv('healthdata.csv') df2 = df.query('`Sleep Disorder`.notnull()')This gives us all rows where the Sleep Disorder column is not null:
Person ID Gender Age ... Heart Rate Daily Steps Sleep Disorder3 4 Male 28.0 ... 85 3000 Sleep Apnea4 5 Male 28.0 ... 85 3000 Sleep Apnea5 6 Male 28.0 ... 85 3000 Insomnia6 7 Male 29.0 ... 82 3500 Insomnia16 17 Female 29.0 ... 80 4000 Sleep Apnea [5 rows x 13 columns]As you can see, our data starts at index 3 because our original source looks like this:
Person ID,Gender,Age,Occupation,Sleep Duration,Quality of Sleep,Physical Activity Level,Stress Level,BMI Category,Blood Pressure,Heart Rate,Daily Steps,Sleep Disorder1,Male,27,Software Engineer,6.1,6,42,6,Overweight,126/83,77,4200,None2,Male,28,Doctor,6.2,6,60,8,Normal,125/80,75,10000,None3,Male,28,Doctor,6.2,6,60,8,Normal,125/80,75,10000,None4,Male,28,Sales Representative,5.9,4,30,8,Obese,140/90,85,3000,Sleep Apnea5,Male,28,Sales Representative,5.9,4,30,8,Obese,140/90,85,3000,Sleep Apnea6,Male,28,Software Engineer,5.9,4,30,8,Obese,140/90,85,3000,Insomnia7,Male,29,Teacher,6.3,6,40,7,Obese,140/90,82,3500,Insomnia8,Male,29,Doctor,7.8,7,75,6,Normal,120/80,70,8000,None9,Male,29,Doctor,7.8,7,75,6,Normal,120/80,70,8000,None10,Male,29,Doctor,7.8,7,75,6,Normal,120/80,70,8000,None...This is a great way to filter out incomplete rows and only work with complete data.
Reversing the Condition
You can also reverse the condition:
df2 = df.query('`Sleep Disorder`.isnull()')This will give us all rows where the Sleep Disorder column is null:
Person ID Gender Age ... Heart Rate Daily Steps Sleep Disorder0 1 Male 27.0 ... 77 4200 NaN1 2 Male 28.0 ... 75 10000 NaN2 3 Male 28.0 ... 75 10000 NaN7 8 Male 29.0 ... 70 8000 NaN8 9 Male 29.0 ... 70 8000 NaN [5 rows x 13 columns]Which can be used to quickly create two datasets - one with complete data and one with incomplete data.
Targeting Index - Not Columns
When using the pandas query, you can also target the index of the rows, not just the columns:
df = pandas.read_csv('healthdata.csv') df2 = df.query('index % 2 == 0')This will give us every second row:
Person ID Gender Age ... Heart Rate Daily Steps Sleep Disorder0 1 Male 27.0 ... 77 4200 NaN2 3 Male 28.0 ... 75 10000 NaN4 5 Male 28.0 ... 85 3000 Sleep Apnea6 7 Male 29.0 ... 82 3500 Insomnia8 9 Male 29.0 ... 70 8000 NaNThis can be achieved by using the index keyword in your query, as it's a special keyword that targets the index of the rows.
Bonus: Different Syntax
As a last example, we wanted to show you another way people do this in tutorials and documentation. This does not use the query method but still filters the data:
df = pandas.read_csv('healthdata.csv') df2 = df[df['Age'] > 30]This will filter the data to only include rows where the Age column is greater than 30:
Person ID Gender Age ... Heart Rate Daily Steps Sleep Disorder32 33 Female 31.0 ... 69 6800 NaN33 34 Male 31.0 ... 72 5000 NaN34 35 Male 31.0 ... 70 8000 NaN35 36 Male 31.0 ... 72 5000 NaN36 37 Male 31.0 ... 72 5000 NaN [5 rows x 13 columns]Performance
One thing to keep in mind is performance. As with everything, method calls have different performance expectations, and you should always test your code to see which method is the fastest for your use case.
In our case, we have tested the performance of the query method and the [] method like this:
main.py
import pandas df = pandas.read_csv('healthdata.csv') df2 = df[df['Age'] > 30]main2.py
import pandas df = pandas.read_csv('healthdata.csv') df2 = df.query('Age > 30')Then, we used the time console command to measure the performance. This is the result:
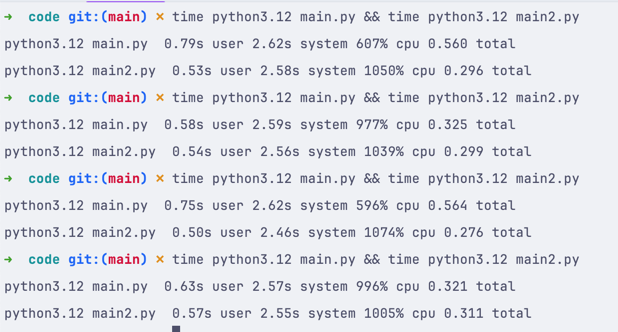
As you can see, the query method is faster in this case. But this might not be the case for your data, so always test your code to see which method/approach is the fastest.
No comments or questions yet...